What is the cost associated with scanning?
How do I prepare my samples?
What size samples can you scan?
What determines resolution?
What type of data is produced, and how do I get it?
Do you accept samples via mail?
Do you do test scans?
What is the cost associated with scanning?
The cost of scanning can be found here. Please note that the XRCT lab prices are subject to change, and you will be charged the rate posted at the time of scanning.
How do I prepare my samples?
Generally speaking sample preparation is not necessary. The ideal geometry for a sample is cylindrical, so if you are cutting smaller samples from larger ones make them cylindrical if possible. For the best results equidimentional samples without sharp edges are best. The sample will need to be mounted to the rotation stage, we have mounts that handle most samples. You should check with us at [email protected] if you have an oddly shaped or large (> 4 cm in diameter) sample.
What size samples can you scan?
There really is no limit on how small the sample can be. The detector we use has an active image area of 23.0 cm x 29.1 cm. The maximum sample size is limited by our detector size and the resolution required.
Resolution
This depends mostly on the sample size and scan geometry. The resolution of our detector is 3889 X 3073 pixels with a 75 µm pixel pitch. The directional tube has a minimum focal spot size of ~ 6 µm, and the transmission tube has a minimum focal spot size of ~ 2 µm. Sample characteristics, such as size and composition will also influence the results. For a general idea of what the resolution (voxel size) will be, divide the maximum dimension of the sample (in mm) by 3889. For example, a sample that has a height of 10 mm the maximum resolution would be 0.0026 mm (2.6 µm). This is just a guideline, there are others factors that must be considered.
Data and data access
Scans done at 16 bit depth generate TIFFs that are 22.75 MB each. If 1000 slices are taken that results in 22.75 GB of raw data, the 3D reconstruction produces more. Some high resolution data (~ 1 µm) can be as large as 350 GB. Slices are provided in the form of 8 bit TIFFs unless another file type is requested. Your folder will contain all of the raw data, the 3D reconstruction, and slices. You will need to download the slice folder, but we encourage you to download everything so you have a backup copy. Refer to the image below for more details.
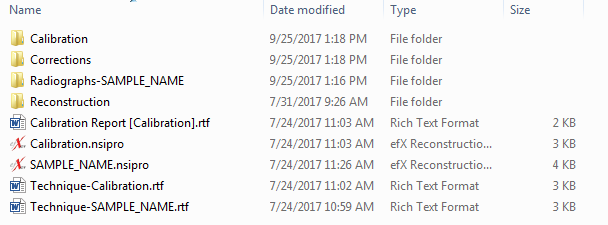
The calibration folder contains the calibration radiographs, the corrections folder contains the images used to correct the display, the radiograph folder contains the sample radiographs, and the reconstruction folder contains the 3D reconstruction. The reconstruction files (.nsidat) are in a proprietary format that you can not use, but we can load them if needed. The .nsipro files are also in a proprietary format but are useful to us if we need to redo the 3D reconstruction. The .rtf files contain information about your scan.
In addition to these folders you will see a folder called 'Slices'. That folder contains the TIFF images you can use for a more quantitative analysis of your sample.
Beginning October 1, 2017 we will begin storing data on Google drive to reduce storage costs. A link to your data will be provided and you will be able to download it using Rclone. Look here for more information about setting up Rclone.
Do you accept samples via mail?
Yes, and in some cases it is desirable to get a sample sent in advance. Return shipping costs must be paid by the customer if the sample is to be mailed back. Our mailing address is: University of Minnesota, Department of Earth Sciences, ATTN: David Birlenbach, 150 John Tate Hall, 116 Church Street SE, Minneapolis, MN 55455.
Do you do test scans?
Yes, but they are not intended for publication. Test scans are mainly for acquiring proof-of-concept images for grant proposals. We will also do a test scan if there is some question about whether or not XRCT will be able to resolve the features you are interested in. Contact [email protected] for more information.